BIO 3170 Final
Which amino acid is usually excluded from alpha helices as they cannot participate in hydrogen bonding?
Alanine
Proline
Histidine
Glycine
What kind of interaction holds together protein secondary structure?
Ionic bonds
Covalent bonds
Hydrogen bonds
London dispersion forces
Van der Waals forces
R groups project ___ from the surface of the alpha helix and determine the chemical nature of the helix
A coiled-coil motif is composed of two alpha helices wound around each other. The seam is stabilized by interactions between heptad ___ and ___ hydrophobic side chains at regular intervals along each strand.
1, 4
2, 3
1, 5
3, 5
The zinc-finger motif is present in many DNA-binding proteins that help regulate what?
Transcription
Translation
Post-transcriptional modification
Post-translational modification
The zinc-finger motif is a 25-residue motif with two invariant __ residues and two invariant __ residues
Thymine
Tryptophan
Cysteine
Proline
Histidine
Leucine
What type of interaction helps to stabilize tertiary protein structure?
Hydrophobic
Hydrophilic
Match the ways to visualize proteins
0%
0
0%
0
0%
0
0%
0
0%
0
0%
0
0%
0
0%
0
The transcription initiation complex is an example of what type of structure?
Secondary structure
Tertiary structure
Quaternary structure
Supramolecular complex
Trypsin is a serine protease whose substrate-binding pocket contains a catalytic site which is composed of which three amino acids?
Serine
Aspartic acid
Leucine
Valine
Histidine
Phenylalanine
Polyubiquitinylation targets proteins for proteasomal degradation. Put its steps in order
Step 4
E1 transfers the Ub molecule to a cysteine residue in E2
Step 2
Enzyme E1 is activated by attachment of a ubiquitin (Ub) molecule
Step 1
E3 adds Ub molecules to the previously added Ub on the target protein
Step 3
Ubiquitin ligase (E3) transfers the E2-bound Ub molecule to the side-chain -NH2 of a lysine residue in a target protein, forming an isopeptide bond
Put the steps of proteolysis in order
4 - Transfer
ATP hydrolysis enables the six-protein (hexameric) ATPase subunits to unfold the substrate
6 - Discharge
Peptide bonds are cleaved
3 - Unfolding
A pore in the hexamer transfers the unfolded protein into the core proteolysis chamber
1 - Recognition
Cleaved peptides (2-24 residues long) are discharged and degraded by soluble proteases
2 - Release
Proteasome 19S cap Ub receptors binds the polyubiquitinylated target
5 - Cleavage
Deubiquitinylase enzyme removes the Ub groups
Match the enzyme to its type of activity
Kinases
Hydrolyze phosphate group off protein
Phosphatases
Transfer terminal phosphate group from ATP to OH group
Transcriptional control is like a switch that can be turned on and off
True
False
Match the type of organism to the mode of gene regulation
Single-celled organism
Genes are regulated to adjust to change in the nutritional and physical environment. Usually produces only the proteins required for survival and proliferation under the particular environmental conditions
Multicellular organism
Genes are regulated to ensure coordination during embryonic development and tissue differentiation
The lac operon encodes 3 enzymes required for the __ of lactose. The trp operon encodes 5 enzymes required for the __ of tryptophan.
Catabolism, anabolism
Catabolism, catabolism
Anabolism, anabolism
Anabolism, catabolism
To initiate transcription, E. coli RNA polymerase must associate with a sigma factor, most commonly sigma __ (number).
The lac repressor binds to the __ of the lac operon when not bound to lactose.
CAP site
Promoter
Operator
LacZ
When lactose is absent, transcription of the lac operon is __.
Promoted
Repressed
E. Coli synthesized cAMP in response to low glucose levels. cAMP binds to a transcriptional activator protein and binds to the __ of the lac operon when complexed with cAMP.
CAP site
Promoter
Operator
LacZ
Match the scenario to the outcome on lac operon expression
Glucose low, cAMP high, lactose absent
Low level of gene expression
Glucose high, cAMP low, lactose absent
No gene expression
Glucose high, cAMP low, lactose present
High level of gene expression
Glucose low, cAMP high, lactose present
No gene expression
The lac repressor always binds to the main operator and one of the two secondary operators.
True
False
The lac operon's default state is __, the trp operon's default state is __.
On, on
On, off
Off, on
Off, off
Enhancers are always found in direct proximity to the gene they regulate.
True
False
Glucorticoid receptor promotes transcription of target genes when glucorticoid hormones binds to the __ activation domain
C-terminal
N-terminal
What are the four major classes of activation (transactivation) domains?
Basic
Glutamine-rich
Proline-rich
Cysteine-rich
Acidic
Isoleucine-rich
What structural motif of DNA binding domains interacts with the major groove of the target DNA?
Alpha helix
Beta sheet
Loop
Turn
Associate the binding domain to the mode of binding.
Leucine-zipper
Heterodimer
C4 zinc-finger
Heterodimer
Basic helix-turn-helix
Homodimer
What is the most common DNA-binding motif?
C2H2 zinc-finger
C4 zinc-finger
Leucine-zipper
Basic helix-turn-helix
CREB (cyclic AMP response element-binding protein) is activated by what?
Acetylation
Phosphorylation
Ubiquitinlyation
Biotinylation
Combinatorial transcription regulation involved interaction of structurally unrelated transcription factors bound to closely spaced binding sites in DNA.
True
False
Which proteins make up the enhanceosome complex of the beta-interferon enhancer?
IRF-3
IRF-7
CJun/AFT-2
P50/p65
HMGI
The ligand-receptor complex functions as a transcription activator.
True
False
Match the functional region of the nuclear receptor to its function.
DNA-binding domain
Contains a hormone-dependent activation domain. In some nuclear receptors, it functions as a repression domain in the absence of a ligand.
N-terminal region
Unique sequence of variable length - 100-500 amino acids. Variable region parts function as activation domains in most nuclear receptors.
Hormone-binding domain
Near the center of the receptor primary sequence. Contains a repeat of the C4 zinc-finger motif.
Put the steps of hormone-dependent gene activation in order.
Step 2
Ligand-binding domain and an additional activation domain at the N-terminus stimulate transcription of target genes
Step 1
DBD binds to response elements
Step 3
It causes a conformational change that releases the chaperone proteins.
Step 4
Hormone complex diffuses through the plasma membrane and binds to the receptor ligand-binding domain
Step 5
Receptor-hormone complex translocates into the nucleus
Response elements contain inverted repeats separated by how many base pairs?
For class I receptors, the hormone interacts with the receptor in the __, while for class II receptors it interacts in the __.
Match the heterodimeric nuclear receptor with its function.
RXR-RAR
Mediates thyroid hormone responses by binding to a direct repeat response element separated by four base pairs
RXR-TR
Mediates vitamin D3 responses by binding to a direct repeat response element separated by three base pairs
RXR-VDR
Mediates retinoic acid responses by binding to a direct repeat reponse element separated by five base pairs
In heterodimeric nuclear receptors, which subunit defines the hormone response and direct repeat binding sites?
RXR subunit
Variable nuclear-receptor subunit
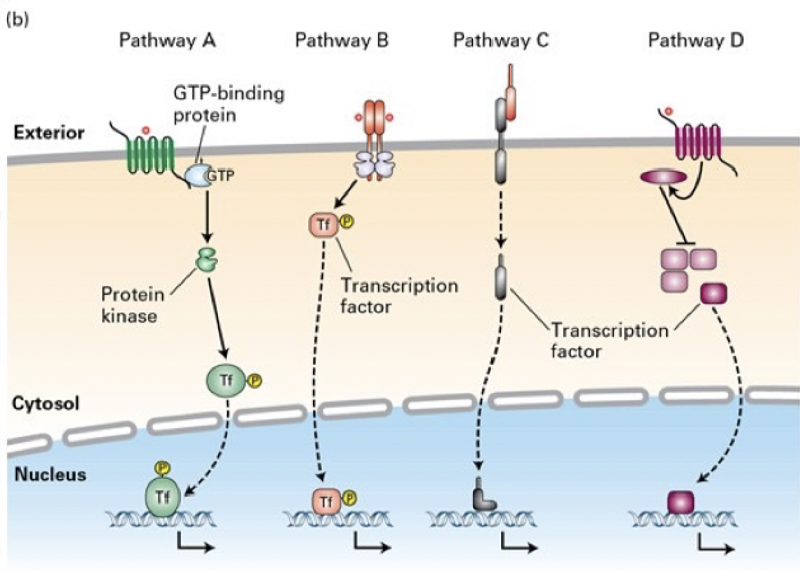
Match the type of cell-surface receptor/signal transduction pathway to its mechanism
Pathway B
Binding of a Delta ligand to the extracellular domain of a member of the Notch family of receptors triggers proteolytin cleavage of the receptor, releasing its cutosolic domain, which moves into the nucleus and regulates gene expression
Pathway D
The cytosolic domains of receptors for the TGF-B family of signaling proteins contain a serine-threonine kinase, which directly phosphorylates and activates a member of the Smad class of transcription factors, unmasking a nuclear localization signal
Pathway A
Many GPCRs (G-protein coupled receptors) activate the heterodimeric GTP-binding protein, ultimately leading to activation of protein kinase A, and phosphorylation and activation of transcription factors such as CREB
Pathway C
Signal transduction pathways activated by binding of members of the Wnt, Hedgehog, or Interleukin 1 (IL-1) families of ligands to their respective receptors lead to ubiquitination and degradation of components of nultiprotein complexes in the cytosol, releasing a transcription factor that then translocates into the nucleus
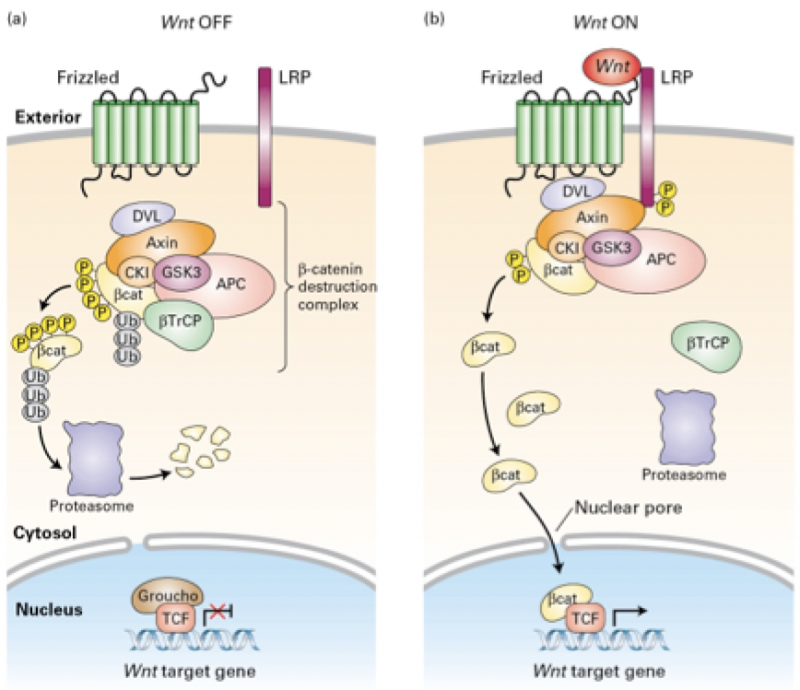
Match the protein to its function in the Wnt signaling pathway (i swear if he asks us about this)
CK1 and GSK3
Phosphorylate beta-catenin at multiple serine and threonine residues
Groucho
Binds to TCF and recruits co-activator proteins to activate expression of Wnt target genes
E3 (betaTrCP)
Binds to phosphorylated LRP, tumor suppressor, component of the destruction complex
Frizzled and LRP
Repressor protein that associates with TCF and inhibits gene transcription
Beta-catenin
Receptor proteins on the cell surface
Axin
Bound to promoters or enhancers of Wnt target genes in the absence of Wnt
Transcription factor TCF
Binds to phosphorylated residues on beta-catenin, leading to ubiquitinylation of beta-catenin and its degradation in proteasomes
How is beta-catenin tagged for degradation in the absence of Wnt?
Acetylation
Ubiquitinylation
Methylation
Phosphorylation
Match the type of gene expression "logic circuit" to its name
0%
0
0%
0
Provides an efficient way to amplify a signal in one direction.
0%
0
0%
0
A single gene encodes a repressor for an entire battery of genes. When this repressor gene is repressed, the battery of genes is expressed.
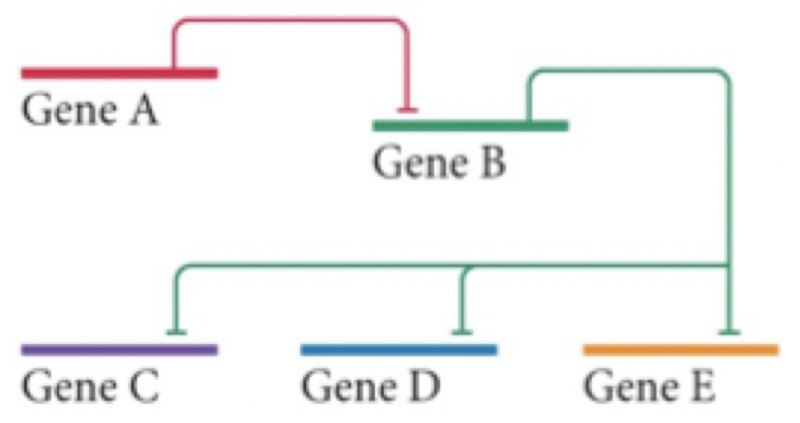
In this gene expression circuit, what will happen is gene A is active?
B not active, C, D, E not active
B not active, C, D, E, active
B active, C, D, E, not active
B active, C, D, E, active
What are the five master transcription factors (Yamanaka factors) that regulate embryonic stem cell pluripotency?
You are able to induce the dedifferentitation of fibroblast cells back into pluripotent stem cells by inserting Yamanaka factors.
True
False
Morphogen gradients help establish what in growing organisms? (multiple answers are correct)
Stem cell orientation
Cell differentiation
Body axes
Orientation of organs
A cut planaria worm is able to regenerate a head and tail in the proper spots due to gradients of which protein?
Wnt
Hn
Notum
Shh
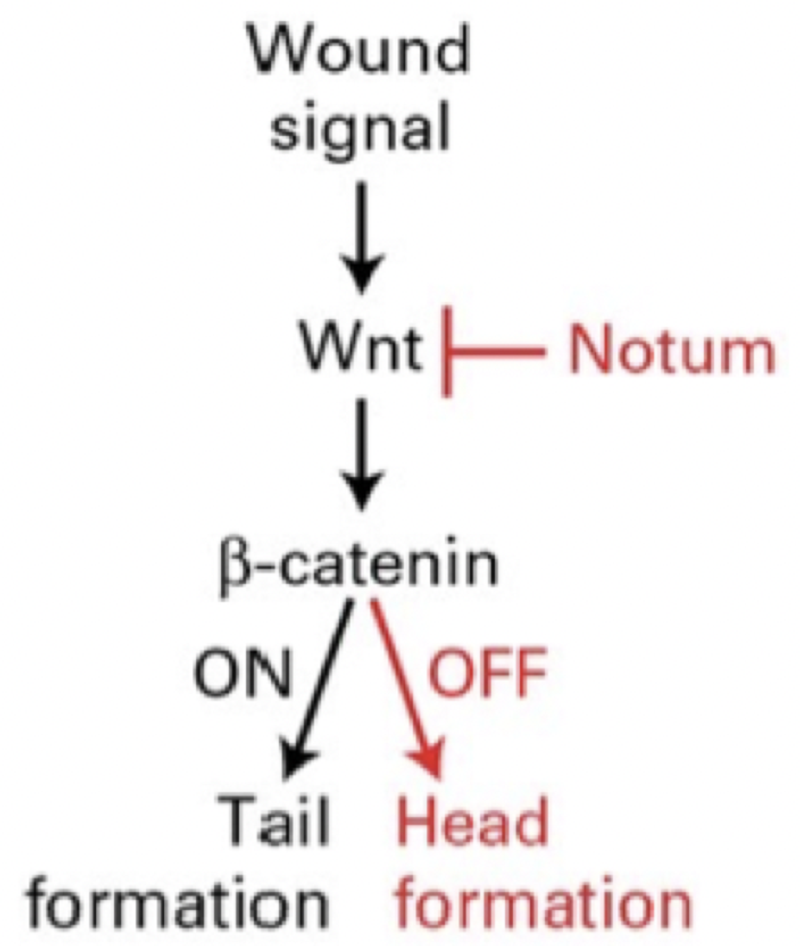
Match the treatment of done on a cut planaria with the result
Inhibitory RNA specific for beta-catenin and notum
Two-tailed planarian
Inhibitory RNA specific for beta-catenin
Two-headed planarian
Inhibitory RNA specific for notum
Two-headed planarian
Veratrum californicum makes cyclopamine, which inhibits __ synthesis, which is needed (Post-Translational Modification) for Hedgehog production and reception.
Dividing nuclei are given information of their position along the axis in flies by the ratio of which two proteins?
Nanog
Wnt
Nanos
Acorn
Sonic
Bicoid
In Drosophila, axis determination genes that activate gap genes come from the mother.
True
False
Match the genes involved in Drosophila axis anterior-posterior formation to their function
Gap genes
Divide the embryo into segment-sized units along the anterior-posterior axis
Maternal axis determination genes
Enable the expression of the pair-rule genes
Segment polarity genes
Form gradients and regions of morphogenic proteins, activate the gap genes
Pair-rule genes
Divides the embryo into regions about two segments wide
In Drosophila embryo, the bicoid protein is most concentration at the __ end.
Anterior
Posterior
MicroRNAs hybridize with which region of the target mRNA?
5' UTR
3' UTR
Promoter
Coding
PolyA tail
5' cap
Match the type of inhibiting RNA to its functions
MiRNA
Hybridize imperfactly with target mRNAs
MiRNA
Causes cleavage of mRNA, triggering its rapid degradation
SiRNA
Repress translation of target mRNAs
MiRNA
More common
SiRNA
Less common
SiRNA
Hybridize perfectly with target mRNA
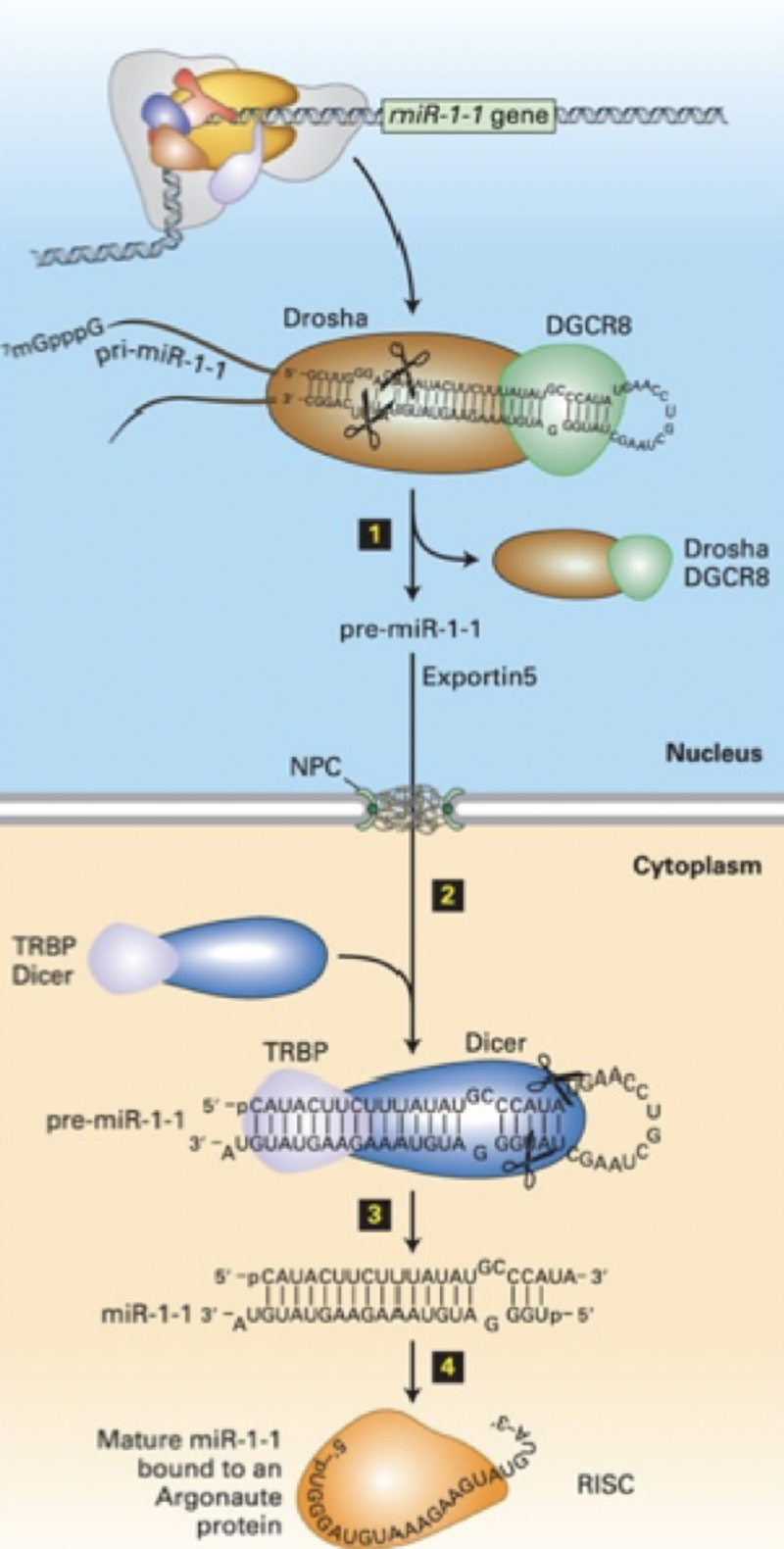
Put the steps of miRNA biogenesis/processing in order
Step 3
Dicer in conjunction with the double-stranded RNA-binding protein TRBP (Loquacious) processes pre-miRNA into a double-stranded miRNA with a two-base single-stranded 3' end
Step 1
Nuclear double-stranded RNA-specific endoribonuclease Drosha and double-stranded RNA-binding protein DGCR8 (Pasha) bind pre-miRNA double strand regions. Drosha cleaves the pre-miRNA.
Step 2
RISC complex incorporates mature miRNA into complex with Argonaute proteins. miRNA-RISC complexes associate with target mRNAs by base pairing between the Argonaute-bound mature miRNA and complementary regions in the 3' UTRs of target mRNAs
Step 4
Exportin 5 - nuclear transporter transports processed pre-miRNA to the cytoplasm
Associate the proteins involved in miRNA biogenesis/processing with their function
RISC complex
Incorporates mature miRNA into complex with Argonaute proteins
TRBP (Loquacious)
Nuclear transporter - transports processed pre-miRNA to the cytoplasm
DGCR8 (Pasha)
Double-strand RNA-binding protein - binds pre-miRNA double strand regions
Dicer
Double-stranded RNA-binding protein - processes pre-miRNA into a double-stranded miRNA
Drosha
Nuclear double-strand RNA-specific endoribonuclease - binds pre-miRNA double strand regions and cleaves it
Exportin 5
Processes pre-miRNA into a double-stranded miRNA
MiRNAs can be used as disease biomarkers
True
False
In bees, miR162a was shown to __ gene expression levels of the cell-signalling pathway regulator TOR
Promote
Inhibit
In bees, what is the main factor that determines formation of worker bees vs. Queen bees?
Location in the hive
Genome
Hatching time
Diet
During early embryogenesis, DNA is heavily methylated
True
False
De novo methylation is mediated by methyltransferase DNMT_, and maintained by DNMT_ following DNA replication.
4,2
3,1
2,3
1,4
When methylation affects CpG islands, methyl-binding proteins trigger a silencing cascade that involved the deacetylation and methylation of histone __.
DNA methylation is heritable
True
False
Which protein ensures methylation patterns are faithfully replicated by scanning DNA and depositing methyl groups on newly synthesized DNA, opposite methyl groups on the old strand?
DNMT3
DNMT1
H3K9
HP1
Match the type of epigenetic inheritance to its definition
Transgenerational Inheritance
Progeny which were not exposed directly to the environmental challenge are nevertheless affected.
Intergenerational Inheritance
The effect is observed in the F1 or F2 progeny of the animals exposed to the environmental trigger. The embryo or its germ cells might be directly exposed to the environmental cue in utero.
The histone code is influenced by environmental factors
True
False
Induction of a gene by a transcription factor can still occur if the target DNA is condensed.
True
False
Histone tails are generally much more extensively __ in euchromatin than in heterochromatin.
This protein contributes to heterochromatin condensation by binding to histone H3 N-terminal tails trimethylated at lysine 9
DNMT1
DNMT3
HP1
HMT
Match the domains/proteins associated with histone deacetylation and hyperacetylation with their function
Gcn5
Interacts with specific upstream activating sequences (UAS) of yeast genes it regulates
DNA-binding domain (deacetylation)
Binds Sin3, a subunit of a multiplrotein complex that includes Rpd3, a histone deacetylase
Rpd3
Hyperacetylates histone N-terminal tails on nucleosomes in the vicinity of the Gcn4-binding site. Decondenses chromatin to facilitate access by the general transcription factors to the TATA box. Activates gene expression.
DNA-binding domain (hyperacetylation)
Interacts with a specific URS1 upstream control element of the genes it regulates
Repression domain
Deacetylates histone N-terminal tails on nucleosomes in the region of the Ume6-binding site. Inhibits general transcription factor binding to the TATA box. Represses gene expression.
Activation domain
Interacts with multiprotein SAGA histone acetylase complex that includes the Gcn5 catalytic subunit
Restriction sites are typically __, and yield fragments with either blunt or sticky ends.
Long
Staggered
Rheumatic
Palindromic
Plasmids replicate jointly with the cell's chromosomal DNA
True
False
How can you differentiate between cells that have successfully taken up your fusion plasmid vs. Cells whose plasmids have re-ligated? (alpha-complementation)
What are the three major steps of PCR amplification?
Initiation
Annealing
Transcription
Denaturing
Extension
Elongation
Put the steps of the CRISPR defensive pathway in order
Step 3
CRISPR RNAs are transcribed from the CRISPR locus
Step 2
A copy of the invading nucleic acid is integrated into the CRISPR locus
Step 1
CRISPR RNAs are incorporated into effector complexes, where the crRNA guides the complex to the invading nucleic acid and the Cas proteins degrade this nucleic acid
What are the two ways the cell repairs target-gene inactivation by the CRISPR-Cas9 system?
Homology-directed repair (HDR)
Repairs the double-strand cleavage using a homologous region in the genome as a template
Nonhomologous end joining (NHEJ)
Repairs the double-strand cleavage, but removes a few bases at the cleavage site
The last universal common ancestor (LUCA) was likely: (check all that apply)
Aerobic
Anaerobic
Hydrophilic
Thermophilic
CO2-fixing
H2-gas dependent
N2-fixing
Heterotrophic
MRNA sequences cannot be modified after transcription.
True
False
{"name":"BIO 3170 Final", "url":"https://www.quiz-maker.com/QPREVIEW","txt":"Which amino acid is usually excluded from alpha helices as they cannot participate in hydrogen bonding?, What kind of interaction holds together protein secondary structure?, R groups project ___ from the surface of the alpha helix and determine the chemical nature of the helix","img":"https://www.quiz-maker.com/3012/CDN/96-4715118/screen-shot-2023-12-10-at-9-40-09-pm.png?sz=1200-00000000000994607153"}
More Quizzes
Simple sentences
1050
How modern are you
420
How Well Do You Know Phoebe?
1890
What power puff girl are you?
320
7th Grade Plant & Animal Cell - Test Your Knowledge
201024012
Test Your Intermediate Computer Skills Now - Free
201081943
Ace Your ABA Knowledge: Free ABA Questions and Answers
201078819
Precalculus Chapter 2 Test Challenge - Ace Your Skills
201089057
Free Electron & Molecular Geometry
201022890
Test Your Skills with a Free Possessive Nouns Assessment
201027273
Which Evil Overlord Are You? Take the Ultimate
201024012
Harry Potter Teachers at Hogwarts: Which One Are You?
201027832
